Multi-Fluorescence Imaging (Optional)
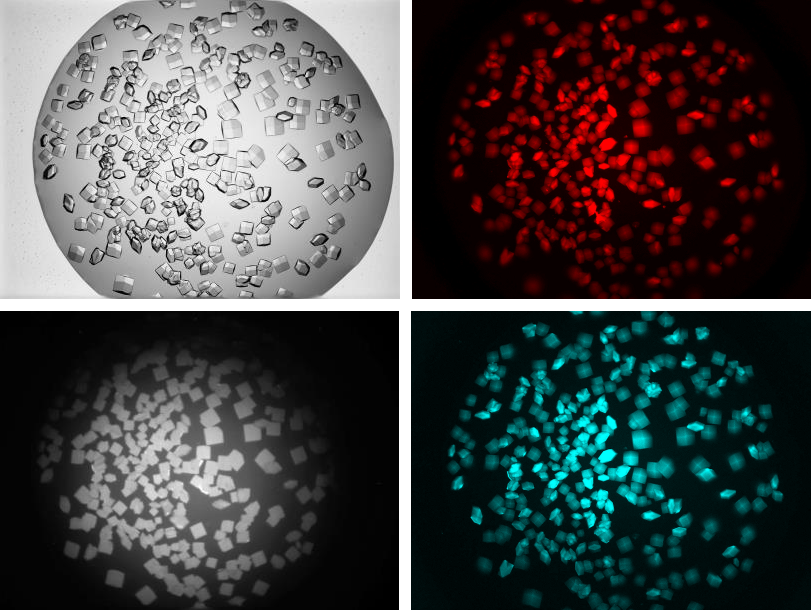
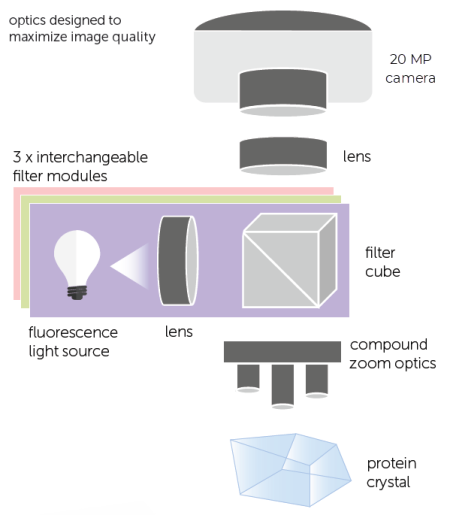
FORMULATRIX’s Multi-Fluorescence Imaging (MFI) technology uses three different light wavelengths, including ultraviolet (UV) and visible fluorescence, to easily detect protein crystals, even those buried under precipitate. MFI includes a full continuous zoom visible imaging optical setup and can be integrated into all ROCK IMAGER 2, 360, and 1000 models.
MFI has three different filter modules that enable you to image the intrinsic UV fluorescence from aromatic amino acids and to detect fluorescence from labeled proteins. Labeling only 0.1% of the proteins in solution with a simple one-step protocol can generate high-contrast fluorescence images of all proteins, including those without tryptophan.
Key Features
- Find hidden protein crystals and distinguish them from salt, even protein with little-to-no-tryptophan.
- Differentiate between crystals of a protein-protein complex and crystals of just a single protein.
- Image faster — 96-well plates can quickly be imaged in about five minutes using visible fluorescence. UV imaging times vary based on tryptophan concentration but average about 10 minutes per plate.
- Use compound zoom optics of 3.3x and 6.6x objectives.
- Optimize image quality and dye-label compatibility through interchangeable filter modules.
How Does MFI Work?
MFI’s optics are optimized for both UV and visible fluorescence imaging to provide high-contrast images. For example, the UV LED and condenser lenses are positioned to maximize the intensity of the UV illumination, boosting signal strength and producing high-quality UV images.
ROCK IMAGER MFI comes with three different filter modules capable of imaging your crystal with different excitation and emission wavelengths. The standard package includes one 290 nm excitation module for UV imaging and two visible fluorescence filter sets for detecting the fluorescence from Texas Red and Fluorescein dyes.
If you would like to image fluorescence from other dyes and install a different filter module, contact support@formulatrix.com.
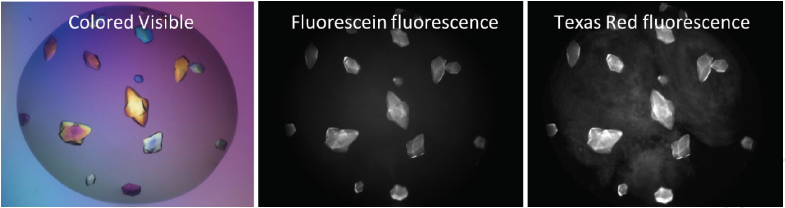
Finding protein-protein complexes
MFI allows you to easily differentiate between crystals of a protein-protein complex and crystals of a single protein.
To find protein-protein complexes:
- Label two suspected proteins or subunits with two different dyes.
- Using the ROCK IMAGER software, image your drop at the two corresponding wavelengths.
Crystals that fluoresce at both wavelengths are protein-protein complexes, whereas those that fluoresce at only one wavelength are of a single protein.
Labeling Protein Sample
To image proteins with the visible fluorescence filter sets, proteins must be intrinsically fluorescent in the visible range (e.g., GFP) or labeled with the appropriate dye.
Proteins can be labeled at any concentration and with any method selected. Many published protocols are available, and care should be taken to use one that is amenable for your specific protein sample. A simple, one-step labeling protocol is suggested below.
To label a protein sample:
Simple 0.1% protein labeling protocol2.
- Prepare a 5 mM stock solution of a succinimidyl ester dye in DMSO.
- Add appropriate amount of dye to protein solution for 0.1% labeling of lysine residues assuming a 1:1 stoichiometric labeling efficiency to amine residues.
- Wait for five minutes, at which point 90% of the dye is bound.
Purification is not required, and samples remain fluorescent for up to 120 days without any known negative impact on crystallization 3,4,5.
Footnotes:
- 1 Images acquired at Maier Lab at Biozentrum Basel.
- 2 Labeling protocol developed by Moritz Hunkeler, Maier Lab at Biozentrum Basel.
- 3 Pusey M., et al. Acta Cryst. (2015) F71, 806-814.
- 4 Watts, D., et al. Acta Cryst. (2010) D66, 901-908.
- 5 Groves, M.R., et al. Acta Cryst. (2006) D63, 526-535.
Related Topics
FORMULATRIX® is proudly powered by WordPress